Life as a PhD student is busy and doesn’t leave much time for other activities, including this blog. So last time, about a month ago, I left you with the question of how the genetic code may have evolved over time.
For decades some scientists have hypothesised that the genetic code evolved by a so-called direct templating mechanism (also known as the stereochemical hypothesis). That is, the strings of ribonucleotides that make up an RNA molecule could physically interact with amino acids, the building blocks of proteins. This interaction would promote the reaction of adjacent amino acids to start forming a longer polypeptide chain. For a review on the different hypotheses see Koonin & Novozhilov (2009).
One of the proponents of the stereochemical hypothesis is Bojan Zagrovic and his research group at the Max F. Perutz Laboratory in Vienna. They have published several papers on this topic and almost a year and a half ago I went to a symposium where Bojan Zagrovic gave a talk on exactly this topic. I wrote about the various presentations I heard there and then several months later a friend I had met during the Cold Spring Harbor Laboratory (CSHL) undergraduate research programme sent me a message saying he had been inspired, by the blog post, to do some research of his own.
In particular, John wanted to investigate whether there was a pattern behind the observed interactions between the amino acids in proteins and the ribonucleotides in RNA. To do this he and Rachel (another student from CSHL) used computational biology approaches to study a large published dataset of protein-RNA complexes. They found that there is a correlation between these physical interactions and the way the genetic code is laid out.
Once these findings had been made they wrote up a draft manuscript, including some figures, which were produced by Grace, a colleague of John’s at Carleton College in the USA. John asked whether I would mind reading the manuscript to give feedback and of course I was happy to do that. We started e-mailing back and forth and decided to extend the computational experiments, and I edited and expanded the text.
The most interesting result was that we could use the knowledge derived solely from the interaction data (blue and red bars) to predict, significantly more accurately than expected by chance (yellow bars), the amino acid sequence of a protein from its mRNA precursor:
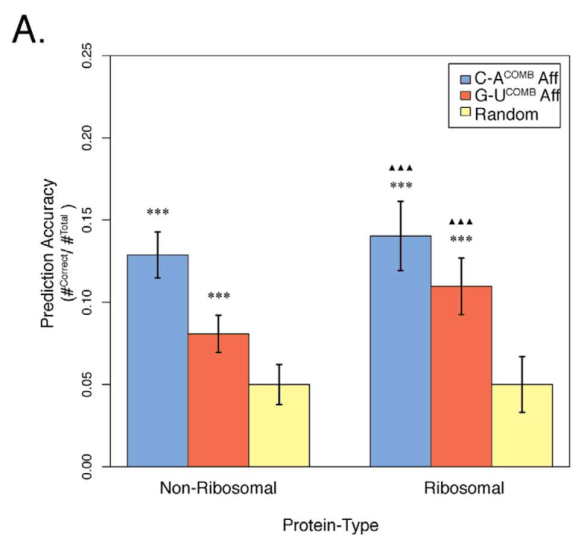
Combining amino acid-nucleobase affinities with mRNA nucleobase content to predict amino acid sequences without universal genetic code. Copied directly from our paper.
In particular, the proteins that form the ribosome – the molecular machine that translates mRNA into protein in modern-day cells – were more accurately predicted than a random protein from our dataset, possibly suggesting that direct interactions between RNA and amino acids led to the formation of the first primitive ribosomes. However, as you can see, the prediction accuracies do not exceed 15% so all results from this paper need to be taken with a pinch of salt; I think the best we can do is say that our results strengthen the stereochemical hypothesis but by no means prove it. [In any case, the scientific method is only good at disproving theories.] Since the journal, Scientific Reports, is an open access journal anyone can read the paper here.
Overall, I am just proud that we managed to publish our work after a long and iterative process, including one revision. All of this was done long-distance via Skype and e-mail. We were all working or studying full-time at the same time and moreover, we did this without the help of a professor/group leader. In fact, none of us even has a PhD (yet).
Lastly, I have noticed a mini-surge in views of my blog posts pertaining to PhD interviews. Clearly the invitations for the next year have been sent out and I hope whoever is reading this is finding it helpful and: good luck!
References:
Cannon JGD, Sherman RM, Wang VMY, Newman GA (2015) Cross-species conservation of complementary amino acid-ribonucleobase interactions and their potential for ribosome-free encoding. Scientific Reports 5: 18054
Hlevnjak M, Zagrovic B (2015) Malleable nature of mRNA-protein compositional complementarity and its functional significance. Nucleic Acids Research 43: 3012-3021
Koonin EV, Novozhilov AS (2009) Origin and evolution of the genetic code: the universal enigma. IUBMB Life 61: 99-111
Polyansky AA, Zagrovic B (2013) Evidence of direct complementary interactions between messenger RNAs and their cognate proteins. Nucleic Acids Research 41: 8434-8443
de Ruiter A, Zagrovic B (2015) Absolute binding-free energies between standard RNA/DNA nucleobases and amino-acid sidechain analogs in different environments. Nucleic Acids Res 43: 708-718
